Research
We develop new computational and experimental tools to study tissue organization in health and disease. Our major focus is studying dynamically remodeling systems such as the ovary and cardiovascular system. Tissue function arises from the coordinated behaviors of cells across time and space. In homeostasis, diverse groups of cells collectively sense, integrate, and respond to local and global signals. How cell circuits integrate combinations of finite signals to regulate diverse outputs while maintaining coherent tissue function is a fundamental question in cell signaling, gene regulation, and physiology. Because our ability to understand is fundamentally limited by what we can measure and model, we work at the interface of cutting-edge genomic technologies (single-cell and spatial methods for tissue biology), perturbations, and computation.
Technology
Perturbations
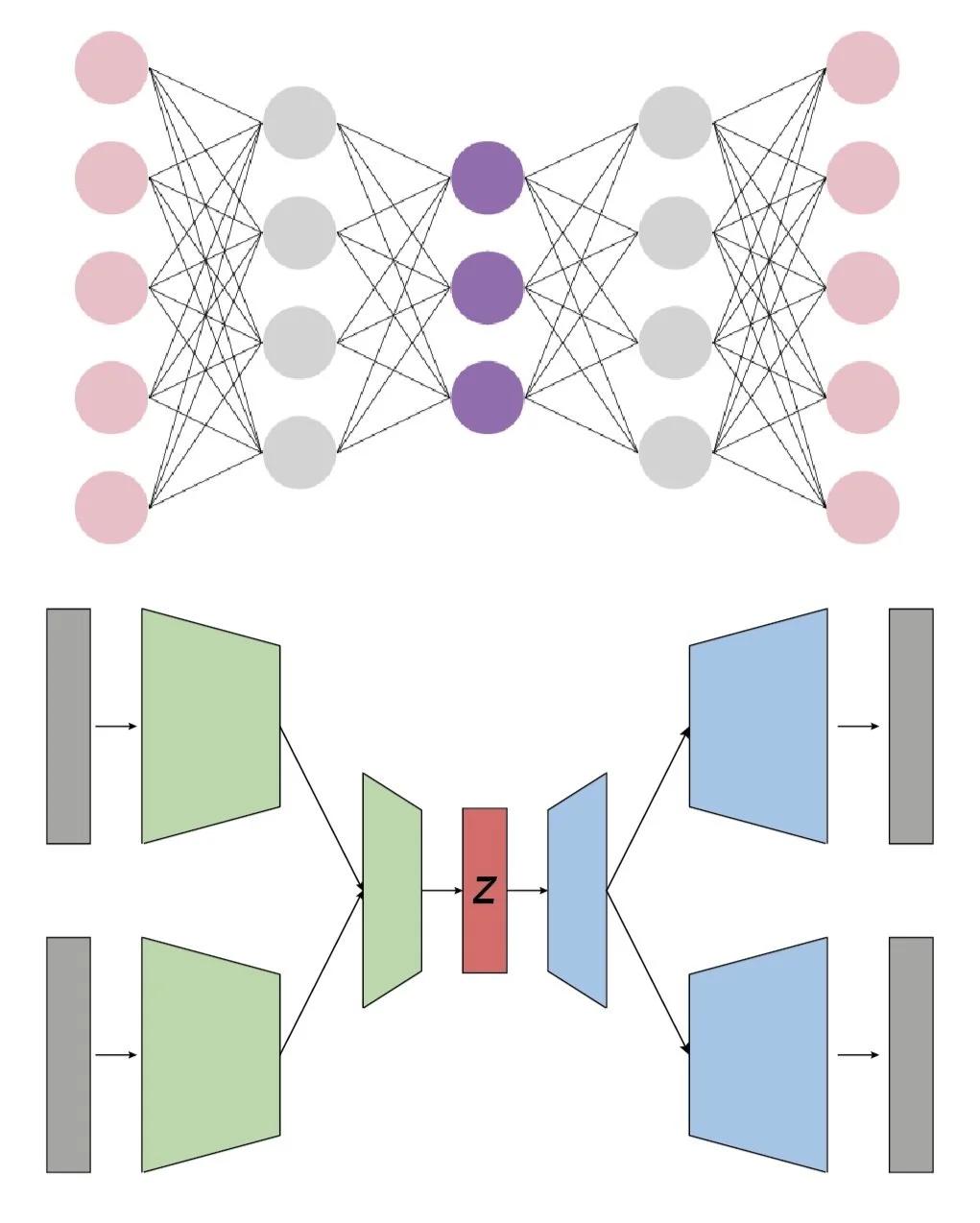
Computation
Current Research Topics
The cardiovascular system is exposed to continuously fluctuating signals that can trigger pathogenic cellular states. We study the molecular drivers of pathogenesis in cardiovascular disease, including coronary artery disease and aortic aneurysms.
Understanding continuous cell state transitions in cardiovascular disease
Dissecting the principles of tissue organization through ovarian remodeling
The ovary is one of the first organs to age. The composition of ovarian cell types, their cellular programs, and interactions within the ovary change dynamically across the estrus/menstrual cycle to control both the reproductive and endocrine functions of the ovary. Throughout this recurrent process, cells undergo continuous phenotypic switching which requires the precise coordination of intra- and inter-cellular signaling. For example, stromal and granulosa cells regulate follicle activation and the delivery of key signaling factors from the vasculature, all of which affect ovulation. Upon ejecting its oocyte, follicles take on a second life by transforming into the corpus luteum, an endocrine structure.
Remarkably, cycling ceases with aging. How these complex cellular interactions change with age is unknown. Because cells in the ovary are structurally organized to form spatially cohesive functional units such as the follicle, cellular functions must be profiled in native tissue contexts. We are applying cutting-edge spatial transcriptomics technologies to understand how diverse cell types contribute to ovarian function and its decline, coupled with machine learning algorithms to model how cells in space coordinate changes in their cellular states over time.
Key questions: What are the functions of immune cells across time and space? How do immune and vascular functions change with age?
Examining the heterogeneity in cellular response to targeted protein degradation therapeutics for hormone-dependent cancers
A central goal in modern medicine is to transform diseased cell states into healthy ones. Degraders are small molecules that harness the ubiquitin system to “delete” target proteins, such as regulatory proteins, thereby altering the genome, epigenome, and transcriptome of a cell. To better understand the molecular mechanisms underlying the varied effects of degraders, including clinical toxicities and cell-type-specific responses, we use single-cell proteogenomic tools like inCITE-seq to simultaneously measure the heterogeneity in drug impact on protein degradation and transcriptomic changes in individual cells.
Novel single-cell and spatial proteogenomics tools
Transforming signals into gene expression is a fundamental cellular computation that drives many biological processes. Building on inCITE-seq (Chung et al. Nature Methods 2021), we are developing advanced single-cell and spatial proteogenomic tools to enable high-dimensional multi-modal measurements that elucidate how signaling patterns drive transcriptional response.